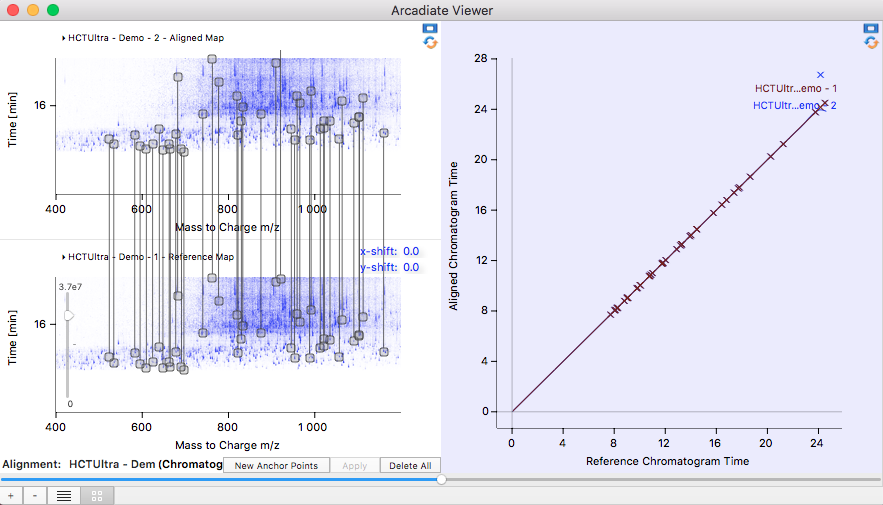
Chromatogram Alignments:
Following a molecule like a peptide over several experiments requires that the chromatograms are aligned to each other. The alignment generates a time axis transformation between the two chromatograms to relate a retention time from one chromatogram to the corresponding retention time of the second chromatogram. Prominent features in the chromatograms are used to calculate the alignment function. A purely computer generated alignment can be incorrect. For this reason Arcadiate offers the possibility to correct automatic alignments or generate them manually altogether.
Chromatogram Normalisations:
The main purpose of Arcadiate is to extract quantitative information from mass spectrometric data sets. The recorded ion intensity or ion volume is not a direct measure of the molecule’s concentration in the sample. The ion volume has to be normalized. There are two widely used normalisation methods: by isotopic labels or by internal standards.
By isotopic labels:
The characteristic of normalizations by isotopic label is that every molecule is measured in comparison to a molecule of nearly identical structure but labelled with heavy isotopes. The kind of label for peptides can be defined by molecular references.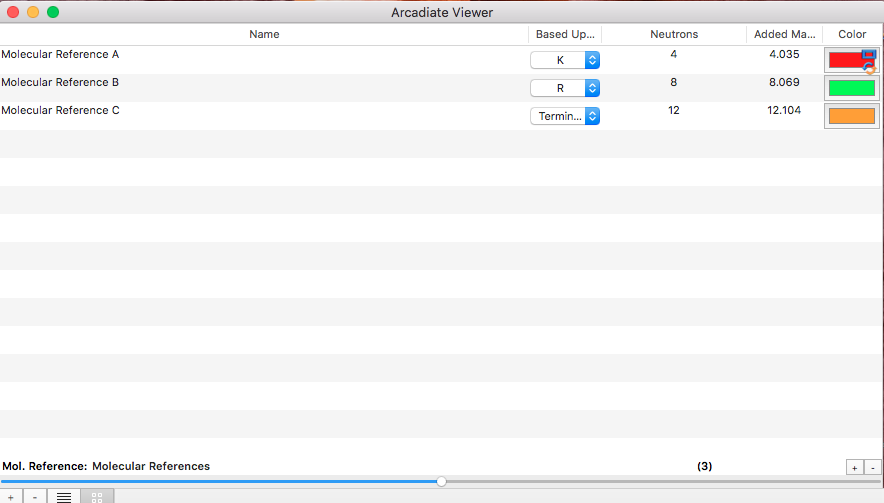
By internal standards:
Internal standard ions are defined by their m/z value, retention time and a specific intensity they should have. In any real experiment the measured ion intensities of the standard ions will deviate from these predefined values. A normalisation function for the entire chromatogram is calculated in such a way that the measured ion intensity multiplied with the actual value of the normalisation function corresponds to the predefined intensity of the standard ion. The normalisation curve is editable to avoid obvious errors in the measurement for instance if one of the normalisation ions has not been detected. 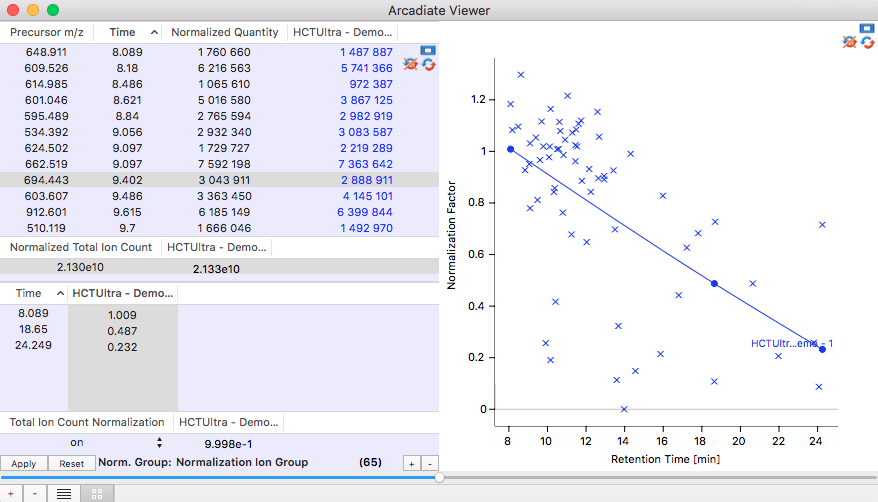
In addition to these peptide-level normalizations total ion count normalizations and protein level normalizations can be used.
Marker Detection in "All Ion Fragmentation" Chromatograms:
"All Ion Fragmentation" chromatograms are acquired by fragmenting entire m/z regions instead of individual precursor ions. Their purpose is to acquire fragment data for close to all ions in a chromatogram. Like this the analysis is much more reproducible than for standard "Data Dependent Acquisition" regimes. One way to use the data is to detect marker ions by the simultaneous presence of their intact m/z value and their fragment ions. Based upon whether such a simultaneous presence can be detected the marker ion is labeled green - when all of its fragment ions are aligned with the intact marker ion , orange - when some but not all of the fragment ions can be aligned with the marker ion - or red when no simultaneous presence can be detected. 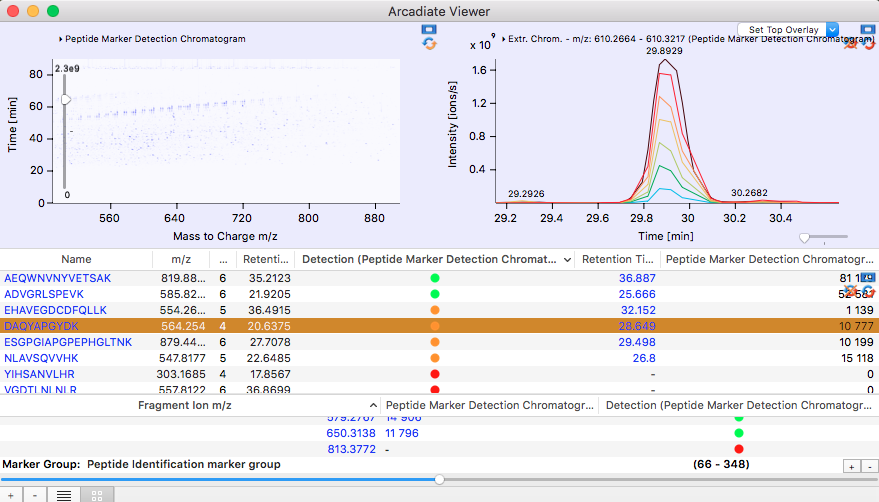
The following page gives a short overview of the inner workings of the application.

